Source (GitHub) | Download notebook
4.1. PopXL and popxl.addons
Introduction
As an alternative to using the ONNX builder to create models, PopXL is an experimental PopART Python module which allows users to hand-craft arbitrary computational graphs, control execution schemes, and optimise the Poplar executable. This provides greater flexibility than is possible using the standard PopART API.
It gives precise control of program size and runtime performance in a high level language (Python). It is useful for large models, as it gives low-level control of:
Subgraph inlining/outlining for optimising program size
Data parallelism for improving throughput
Remote Variables for streaming/storing data remotely to optimise memory requirements of the model
Familiarity with the basic PopXL concepts will be helpful, but not essential:
Intermediate representation (IR)
Tensors (variable, constant and intermediate)
Graphs (main and subgraph)
Input and output streams
Sessions
popxl.addons includes common usage patterns of PopXL. It simplifies the process of building and training a model while keeping control of execution and optimisation strategies.
Once you’ve finished this tutorial, you will:
Be familiar with
addonsbasic concepts and understand relations between them:ModuleNamedTensorsVariableFactoryandNamedVariableFactoriesGraphWithNamedArgsBoundGraphTransforms
Understand what outlining is and write an outlined model.
Use
addonsautodifftransform to obtain the backward graph for your model.Write a simple training + test program, copying trained weights from the training session to the test session.
Requirements
Install a Poplar SDK (version 2.6 or later) and source the enable.sh scripts for both PopART and Poplar as described in the Getting Started guide for your IPU system.
Install system dependencies:
apt-get install -y $(< required_apt_packages.txt)Create a Python virtual environment:
python3 -m venv <virtual_env>.Activate the virtual environment:
. <virtual_env>/bin/activate.Update
pip:pip3 install --upgrade pipInstall requirements
pip3 install -r requirements.txt(this will also install popxl.addons).
python3 -m venv virtual_env
. virtual_env/bin/activate
pip3 install --upgrade pip
pip3 install -r requirements.txt
To run the Jupyter notebook version of this tutorial:
Install a Poplar SDK (version 2.6 or later) and source the enable.sh scripts for both PopART and Poplar as described in the Getting Started guide for your IPU system
Create a virtual environment
In the same virtual environment, install the Jupyter notebook server:
python -m pip install jupyterLaunch a Jupyter Server on a specific port:
jupyter-notebook --no-browser --port <port number>. Be sure to be in the virtual environment.Connect via SSH to your remote machine, forwarding your chosen port:
ssh -NL <port number>:localhost:<port number> <your username>@<remote machine>
For more details about this process, or if you need troubleshooting, see our guide on using IPUs from Jupyter notebooks.
If using VS Code, Intellisense can help you understand the tutorial code. It will show function and class descriptions when hovering over their names and lets you easily jump to their definitions. Consult the VSCode setup guide to use Intellisense for this tutorial.
Basic concepts
The following are basic concepts of popxl.addons:
Module: a module is the base class to build layers and models. It is similar to a PyTorch Module or a Keras Layer. It allows you to create a graph with state, which means with internal parameters (such as weights). Graphs can be called from multiple places in parent graphs, which reduces code duplication.NamedTensors: aDotTreecollection ofpopxl.Tensor, which is basically a dictionary between names and tensors.DotTreeare used inaddonsfor all collections of named objects. They are very useful to group objects in namespaces.VariableFactoryandNamedVariableFactories(aDotTreecollection of factories): factories are used to delay variable instantiation to the main graph.GraphWithNamedArgs: it is a container of apopxl.GraphandNamedTensors.NamedTensorsare the graph named inputs. When aGraphWithNamedArgsis created withModule.create_graph(), factories for each of the input tensors are instantiated as well.transformsare functions that directly modify the graph, generating a new one. An example is theaddons.autodifftransform, which, given a graph, produces the corresponding backward graph.BoundGraph: it is a container of apopxl.Graphand aTensorMapof bound Tensor inputs, which are automatically provided as inputs when the graph is called.
The figure below shows how these concepts are related in a typical popxl.addons workflow:
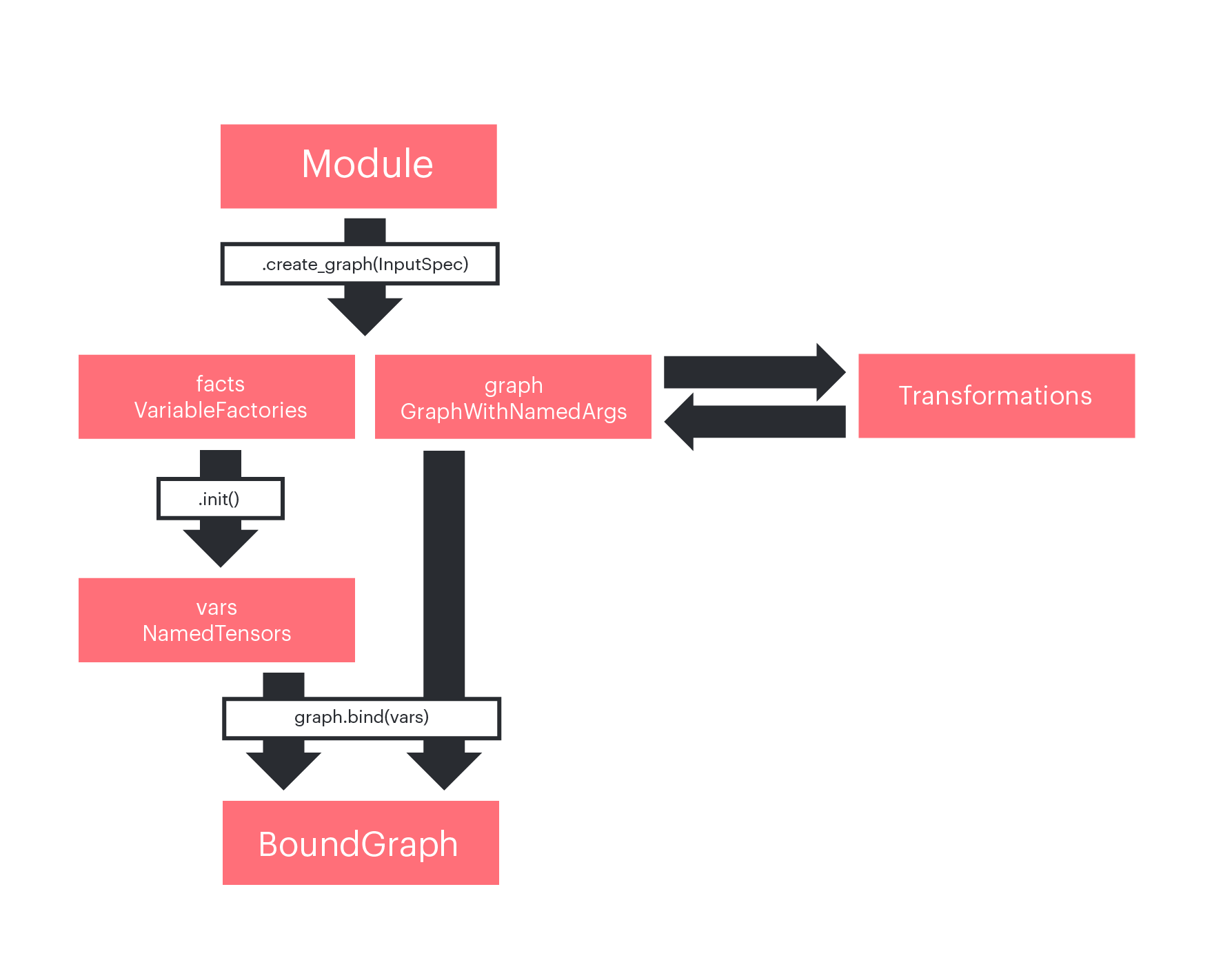
Figure 1: Workflow in popxl.addons.
A simple example
Imports
First let’s import all the packages we are going to need, including popxl and
popxl_addons:
from tqdm import tqdm
import torchvision
import torch
import numpy as np
from typing import Mapping, Optional
from functools import partial
np.random.seed(42)
import popxl.ops as ops
import popxl_addons as addons
import popxl
Defining a Linear Module
Below is an example of a linear layer implemented by subclassing
addons.Module:
class Linear(addons.Module):
def __init__(self, out_features: int, bias: bool = True):
super().__init__()
self.out_features = out_features
self.bias = bias
def build(self, x: popxl.Tensor) -> popxl.Tensor:
# add a state variable to the module
w = self.add_variable_input(
"weight",
partial(np.random.normal, 0, 0.02, (x.shape[-1], self.out_features)),
x.dtype,
)
y = x @ w
if self.bias:
# add a state variable to the module
b = self.add_variable_input("bias", partial(np.zeros, y.shape[-1]), x.dtype)
y = y + b
return y
Each Module needs to implement a build method. Here you actually define the
graph, specifying inputs, operations and outputs (the return values).
Inputs are added in two different ways:
named inputs (the parameters of the model, its state) are added via the
add_variable_inputmethod (wandbabove).All tensor arguments of the
buildmethod (xin the example above) become inputs of the graph.
PopXL variables (the parameters of the model) can only live in the main graph
which means state tensor variables cannot be instantiated directly in the
subgraph; their creation needs to take place in the main graph. The
add_variable_input method creates a named local placeholder (a local tensor)
and a corresponding VariableFactory. As we will see below, the factory is used
to instantiate a variable in the main graph and then bind it to the named input
in the subgraph.
Creating a graph from a Module
Before we construct any graphs we need to define a popxl.Ir instance which
will hold the main graph. This will represent your fully compiled program.
ir = popxl.Ir(replication=1)
Inside the IR main graph we construct the Linear graph as follows:
with ir.main_graph:
# create a variable factory and a graph from the module
facts, linear_graph = Linear(32).create_graph(
popxl.TensorSpec((2, 4), popxl.float32)
)
print("factories: \n", facts)
print("\n graph: \n", linear_graph.print_schedule())
# since we are in the main graph, we can instantiate variables using the factories
variables = facts.init()
print("\n variables: \n", variables)
# and bind our graph to these variables that live in the main graph.
bound_graph = linear_graph.bind(variables)
# the bound graph can be called providing only unnamed inputs x
input_data = np.asarray(np.random.rand(2, 4)).astype(np.float32)
input_tensor = popxl.variable(input_data, name="x")
out = bound_graph.call(input_tensor)
factories:
{'weight': <popxl_addons.variable_factory.VariableFactory object at 0x7feaf5cb1160>, 'bias': <popxl_addons.variable_factory.VariableFactory object at 0x7fe851acee10>}
graph:
Graph : Linear_subgraph(0)
(%1, weight=%2, bias=%3) -> (%5) {
MatMul.100 (%1 [(2, 4) float32], %2 [(4, 32) float32]) -> (%4 [(2, 32) float32])
Add.101 (%4 [(2, 32) float32], %3 [(32,) float32]) -> (%5 [(2, 32) float32])
}
variables:
{'bias': Tensor[bias popxl.dtypes.float32 (32,)], 'weight': Tensor[weight popxl.dtypes.float32 (4, 32)]}
The create_graph method requires a TensorSpec object or a Tensor object
for each of the unnamed inputs (x) and returns a GraphWithNamedArgs and
NamedVariableFactories for the named inputs.
The GraphWithNamedArgs gathers together a popxl.Graph and tensors
(GraphWithNamedArgs.args) which correspond to the named inputs we added
earlier with add_variable_input when we defined the Linear Module.
Also, the NamedVariableFactories collection contains a factory for each of
those tensors. Instantiating a variable in the main graph is optional, provided
you feed these inputs at the call site. for example linear_graph.call(x,w,b)
Once you have instantiated variables using these factories, you can bind the
graph to them using bound_graph = linear_graph.bind(variables).
The resulting BoundGraph is effectively a graph with an internal state, which
can be called from the main graph with:
outputs = bound_graph.call(x)
Finally, we specify the properties of the IR and create a session object to run it.
ir.num_host_transfers = 1
# Create a session to run your IR
session = popxl.Session(ir, "ipu_hw")
# Run the program
with session:
session.run()
Summary and concepts in practice
In summary, the basic steps needed to build and run a model in popxl.addons are the following:
Subclass
addons.Moduleto create your layers and models in an object-oriented fashion.Initialise an IR as
irwhich represents your full compiled program.In the
ir.main_graph()context, generate the computational graph and the variable factories associated with your module with theModulecreate_graphmethod.In the
ir.main_graph()context, instantiate actual variables with theNamedVariableFactoryinitmethod.In the
ir.main_graph()context, bind the computational graph to the variables using thebindmethod ofGraphWithNamedArgs.In the
ir.main_graph()context, call the bound graph providing only the inputs.Specify the properties of the IR and create a
popxl.Sessionto run your IR.Run the program.
Multiple bound graphs
The same subgraph can be bound to different sets of parameters, resulting in different bound graphs. You can think to a bound graph as a container of a graph object and a set of variables. Two different bound graphs can reference to the same graph, but have different variables.
ir = popxl.Ir()
with ir.main_graph:
facts, linear_graph = Linear(32).create_graph(
popxl.TensorSpec((2, 4), popxl.float32)
)
variables1 = facts.init()
variables2 = facts.init()
# the same subgraph can be bound to different variables,
# generating distinct bound graph objects.
bound_graph1 = linear_graph.bind(variables1)
bound_graph2 = linear_graph.bind(variables2)
print("\n two different bound graphs: \n", bound_graph1, bound_graph2)
input_data = np.asarray(np.random.rand(2, 4)).astype(np.float32)
input_tensor = popxl.variable(input_data, name="x")
out = bound_graph1.call(input_tensor)
two different bound graphs:
<popxl_addons.graph.BoundGraph object at 0x7fe851b717b8> <popxl_addons.graph.BoundGraph object at 0x7fe851c3b4a8>
Both bound graphs refer to the same computational graph, hence code is reused on the IPU.
What actually happens is that bound_graph1.call becomes a call operation in
the main graph with the following arguments:
call(graph, x, inputs_dict=variables_dict1)
where variables_dict1 is a dictionary between the local tensors in the graph
and the corresponding variables in the main graph, created using the factories.
Calling bound_graph2, which is the same graph but bound to different
parameters, results in:
call(graph, x, inputs_dict=variables_dict2)
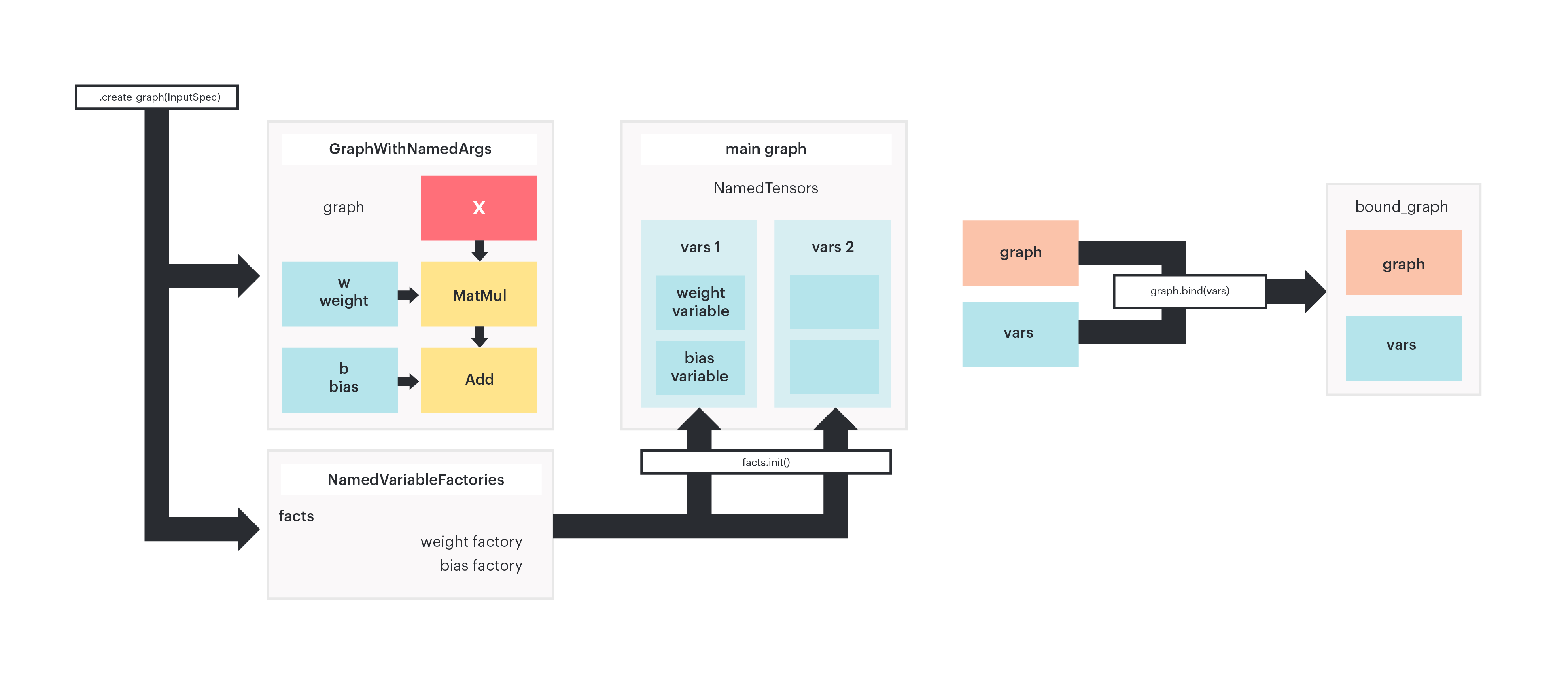
Figure 2: The module.create_graph method generates a graph and named
variable factories. Calling .init() on the variable factories instantiates
actual variable tensors in the main graph. The same subgraph can be bound to
different sets of variables, generating different bound graphs.
Nested Modules and Outlining
Outlining is the opposite of inlining:
Inlining, which is generally the default behaviour on the IPU means that if a module is used more than once in a model, then a copy will be made of the code on the IPU for each use. This has the effect of better performance, because we remove some of the overhead of calling out to a function, but will use more always-live memory due to code duplication.
Outlining, conversely requires some explicit PopXL code. Code will only exist once on each IPU and could be called from multiple parent models. This has the effect of reducing total memory requirements at the expense of some performance.
Further information on graph outlining can be found in our memory performance optimisation guide.
These concepts becomes important when we start building more complicated models
using the
Module
api, combining and nesting several modules. To see why, let’s build a simple
linear model using several Linear layers.
class Net(addons.Module):
def __init__(self, cache: Optional[addons.GraphCache] = None):
super().__init__(cache=cache)
self.fc1 = Linear(512)
self.fc2 = Linear(512)
self.fc3 = Linear(512)
self.fc4 = Linear(10)
def build(self, x: popxl.Tensor):
x = x.reshape((-1, 28 * 28))
x = ops.gelu(self.fc1(x))
x = ops.gelu(self.fc2(x))
x = ops.gelu(self.fc3(x))
x = self.fc4(x)
return x
ir = popxl.Ir()
main = ir.main_graph
with main:
facts, net_graph = Net().create_graph(popxl.TensorSpec((28, 28), popxl.float32))
print(facts.to_dict().keys())
print("\n", net_graph.print_schedule())
dict_keys(['fc1.weight', 'fc1.bias', 'fc2.weight', 'fc2.bias', 'fc3.weight', 'fc3.bias', 'fc4.weight', 'fc4.bias'])
Graph : Net_subgraph(0)
(%1, fc1.weight=%2, fc1.bias=%3, fc2.weight=%4, fc2.bias=%5, fc3.weight=%6, fc3.bias=%7, fc4.weight=%8, fc4.bias=%9) -> (%21) {
Reshape.100 (%1 [(28, 28) float32]) -> (%10 [(1, 784) float32])
MatMul.101 (%10 [(1, 784) float32], %2 [(784, 512) float32]) -> (%11 [(1, 512) float32])
Add.102 (%11 [(1, 512) float32], %3 [(512,) float32]) -> (%12 [(1, 512) float32])
Gelu.103 (%12 [(1, 512) float32]) -> (%13 [(1, 512) float32])
MatMul.104 (%13 [(1, 512) float32], %4 [(512, 512) float32]) -> (%14 [(1, 512) float32])
Add.105 (%14 [(1, 512) float32], %5 [(512,) float32]) -> (%15 [(1, 512) float32])
Gelu.106 (%15 [(1, 512) float32]) -> (%16 [(1, 512) float32])
MatMul.107 (%16 [(1, 512) float32], %6 [(512, 512) float32]) -> (%17 [(1, 512) float32])
Add.108 (%17 [(1, 512) float32], %7 [(512,) float32]) -> (%18 [(1, 512) float32])
Gelu.109 (%18 [(1, 512) float32]) -> (%19 [(1, 512) float32])
MatMul.110 (%19 [(1, 512) float32], %8 [(512, 10) float32]) -> (%20 [(1, 10) float32])
Add.111 (%20 [(1, 10) float32], %9 [(10,) float32]) -> (%21 [(1, 10) float32])
}
You can see from the output of print_schedule() that nested modules lead to
inlined code: the nodes are repeated for each layer, even if they are
identical. For example here fc2 and fc3 have the exact same graph. If you
want to achieve better code reuse you can manually outline the graph by
explicitly inserting call operations.
To implement outlining using the Module class, you need to:
Create the graph you want to outline with
factories, shared_graph = module.create_graph().Generate different named input tensors (different local placeholders) for each layer. To do this, you can use the
module.add_variable_inputs(name, factories)method. Every time you call this function, you create distinct local tensors. Moreover, you add factories for these tensors.Bind the graph to each set of local tensors, obtaining a different
BoundGraphfor each layer.Add call operations to the bound graphs.
When you call factories.init() in the main context you generate variables for
all the local tensors. When you finally bind the graph, the local tensors are
bound to the main variables. Since the shared_graph is bound to the local
tensors, it is effectively bound to them too.
Below is an outlined version of the network. You can see in the graph that the
fc2 and fc3 blocks have been replaced by call operations.
class NetOutlined(addons.Module):
def __init__(self, cache: Optional[addons.GraphCache] = None):
super().__init__(cache=cache)
# first and last layer are not reused
self.fc1 = Linear(512)
self.fc4 = Linear(10)
def build(self, x: popxl.Tensor):
x = x.reshape((-1, 28 * 28))
x = ops.gelu(self.fc1(x))
# create a single subgraph to be used both for fc2 and fc3
# create variable factories and subgraph
facts, subgraph = Linear(512).create_graph(x)
# generate specific named inputs for fc2
named_tensors_0 = self.add_variable_inputs("fc2", facts)
# fc2 is a bound graph using the shared, single subgraph and custom params
fc2 = subgraph.bind(named_tensors_0)
# generate specific named inputs for fc3
named_tensors_1 = self.add_variable_inputs("fc3", facts)
# fc3 is a bound graph using the shared, single subgraph and custom params
fc3 = subgraph.bind(named_tensors_1)
(x,) = fc2.call(x)
x = ops.gelu(x)
(x,) = fc3.call(x)
x = ops.gelu(x)
x = self.fc4(x)
return x
ir = popxl.Ir()
with ir.main_graph:
args, net_graph = NetOutlined().create_graph(
popxl.TensorSpec((28, 28), popxl.float32)
)
print(args.to_dict().keys())
print("\n", net_graph.print_schedule())
dict_keys(['fc1.weight', 'fc1.bias', 'fc4.weight', 'fc4.bias', 'fc2.weight', 'fc2.bias', 'fc3.weight', 'fc3.bias'])
Graph : NetOutlined_subgraph(0)
(%1, fc1.weight=%2, fc1.bias=%3, fc2.weight=%4, fc2.bias=%5, fc3.weight=%6, fc3.bias=%7, fc4.weight=%8, fc4.bias=%9) -> (%19) {
Reshape.100 (%1 [(28, 28) float32]) -> (%10 [(1, 784) float32])
MatMul.101 (%10 [(1, 784) float32], %2 [(784, 512) float32]) -> (%11 [(1, 512) float32])
Add.102 (%11 [(1, 512) float32], %3 [(512,) float32]) -> (%12 [(1, 512) float32])
Gelu.103 (%12 [(1, 512) float32]) -> (%13 [(1, 512) float32])
Call.106(Linear_subgraph(1)) (%13 [(1, 512) float32], %4 [(512, 512) float32], %5 [(512,) float32]) -> (%14 [(1, 512) float32])
Gelu.107 (%14 [(1, 512) float32]) -> (%15 [(1, 512) float32])
Call.108(Linear_subgraph(1)) (%15 [(1, 512) float32], %6 [(512, 512) float32], %7 [(512,) float32]) -> (%16 [(1, 512) float32])
Gelu.109 (%16 [(1, 512) float32]) -> (%17 [(1, 512) float32])
MatMul.110 (%17 [(1, 512) float32], %8 [(512, 10) float32]) -> (%18 [(1, 10) float32])
Add.111 (%18 [(1, 10) float32], %9 [(10,) float32]) -> (%19 [(1, 10) float32])
}
DotTree example
The following code block is a simple example to help you get familiar with the
DotTree syntax used by popxl.addons.
ir = popxl.Ir(replication=1)
with ir.main_graph:
facts, linear_graph = Linear(32).create_graph(
popxl.TensorSpec((2, 4), popxl.float32)
)
variables = facts.init()
variables2 = facts.init()
print("----- DotTree functionalities -----\n")
collection = addons.NamedTensors() # empty collection
collection.insert("layer1", variables) # add a new key with the insert method
collection.insert("layer2", variables2)
print(
"A nested collection: the nested structure appear from the dot structure of names"
)
print(collection, "\n") # nested structure
print("Each leaf can be accessed with dot syntax: collection.layer1")
print(
collection.layer1
) # each leaf node in the tree can be accessed with dot syntax
collection_dict = collection.to_dict() # convert the collection to dictionary
nt = addons.NamedTensors.from_dict(
{"x": input_tensor}
) # create the collection from a dictionary
nt_b = addons.NamedTensors.from_dict({"b": variables.bias})
nt.update(
nt_b
) # update the collection with another collection, keys should not repeat
# print("\n",nt)
names = ["new_bias_name", "new_weight_name"]
nt_from_lists = addons.NamedTensors.pack(
names, variables.tensors
) # create the collection from a list
names, tensors = nt_from_lists.unpack()
# print("\n",nt_from_lists)
print("\n")
# ----- NamedTensors specific -----
print("----- NamedTensors -----\n")
print(collection.named_tensors, "\n") # same as to_dict, specific of NamedTensors
print("Get only tensors, sorted by name")
print(collection.tensors) # returns tensors, sorted by names
----- DotTree functionalities -----
A nested collection: the nested structure appear from the dot structure of names
{'layer1.bias': Tensor[bias popxl.dtypes.float32 (32,)], 'layer1.weight': Tensor[weight popxl.dtypes.float32 (4, 32)], 'layer2.bias': Tensor[bias__t0 popxl.dtypes.float32 (32,)], 'layer2.weight': Tensor[weight__t1 popxl.dtypes.float32 (4, 32)]}
Each leaf can be accessed with dot syntax: collection.layer1
{'bias': Tensor[bias popxl.dtypes.float32 (32,)], 'weight': Tensor[weight popxl.dtypes.float32 (4, 32)]}
----- NamedTensors -----
{'layer1.bias': Tensor[bias popxl.dtypes.float32 (32,)], 'layer1.weight': Tensor[weight popxl.dtypes.float32 (4, 32)], 'layer2.bias': Tensor[bias__t0 popxl.dtypes.float32 (32,)], 'layer2.weight': Tensor[weight__t1 popxl.dtypes.float32 (4, 32)]}
Get only tensors, sorted by name
(Tensor[bias popxl.dtypes.float32 (32,)], Tensor[bias__t0 popxl.dtypes.float32 (32,)], Tensor[weight popxl.dtypes.float32 (4, 32)], Tensor[weight__t1 popxl.dtypes.float32 (4, 32)])
You can experiment with the above code to explore the api before going through the full MNIST program.
MNIST
Load dataset
We are now ready to build the full program to train and validate a linear model on the MNIST dataset.
First of all, we need to load the dataset. We are going to use a PyTorch DataLoader. Data is normalised using the mean and std deviation of the dataset.
def get_mnist_data(test_batch_size: int, batch_size: int):
training_data = torch.utils.data.DataLoader(
torchvision.datasets.MNIST(
"~/.torch/datasets",
train=True,
download=True,
transform=torchvision.transforms.Compose(
[
torchvision.transforms.ToTensor(),
# mean and std computed on the training set.
torchvision.transforms.Normalize((0.1307,), (0.3081,)),
]
),
),
batch_size=batch_size,
shuffle=True,
drop_last=True,
)
validation_data = torch.utils.data.DataLoader(
torchvision.datasets.MNIST(
"~/.torch/datasets",
train=False,
download=True,
transform=torchvision.transforms.Compose(
[
torchvision.transforms.ToTensor(),
torchvision.transforms.Normalize((0.1307,), (0.3081,)),
]
),
),
batch_size=test_batch_size,
shuffle=True,
drop_last=True,
)
return training_data, validation_data
Defining the Training step
Now that we have a dataset and a model we are ready to construct the training program.
First of all let’s define a function to build the training program.
def train_program(batch_size, device, learning_rate):
ir = popxl.Ir(replication=1)
with ir.main_graph:
# Create input streams from host to device
img_stream = popxl.h2d_stream((batch_size, 28, 28), popxl.float32, "image")
img_t = ops.host_load(img_stream) # load data
label_stream = popxl.h2d_stream((batch_size,), popxl.int32, "labels")
labels = ops.host_load(label_stream, "labels")
# Create forward graph
facts, fwd_graph = Net().create_graph(img_t)
# Create backward graph via autodiff transform
bwd_graph = addons.autodiff(fwd_graph)
# Initialise variables (weights)
variables = facts.init()
# Call the forward graph with call_with_info because we want to retrieve
# information from the call site
fwd_info = fwd_graph.bind(variables).call_with_info(img_t)
x = fwd_info.outputs[0] # forward output
# Compute loss and starting gradient for backpropagation
loss, dx = addons.ops.cross_entropy_with_grad(x, labels)
# Setup a stream to retrieve loss values from the host
loss_stream = popxl.d2h_stream(loss.shape, loss.dtype, "loss")
ops.host_store(loss_stream, loss)
# retrieve activations from the forward graph
activations = bwd_graph.grad_graph_info.inputs_dict(fwd_info)
# call the backward graph providing the starting value for backpropagation and activations
bwd_info = bwd_graph.call_with_info(dx, args=activations)
# Optimiser: get a mapping between forward tensors and corresponding
# gradients and use it to update each tensor
grads_dict = bwd_graph.grad_graph_info.fwd_parent_ins_to_grad_parent_outs(
fwd_info, bwd_info
)
for t in variables.tensors:
ops.scaled_add_(t, grads_dict[t], b=-learning_rate)
ir.num_host_transfers = 1
return popxl.Session(ir, device), [img_stream, label_stream], variables, loss_stream
We need to specify the replication_factor for the IR before constructing the
program, because some operations need to know the number of IPUs involved.
Inside the main graph context of the IR, we construct
streams
to transfer input data from the host to the device (popxl.h2d_stream) and we
load data to the device (popxl.host_load).
Then, we create two graphs: one for the forward pass and one for the backward
pass. The latter can be obtained from the forward graph applying a
transform, which is a way of making changes at the graph level. The
addons.autodiff transform is the one we need. It is basically
popxl.autodiff
with some additional patterns.
We instantiate the weights of the network (variables = facts.init()), bind the
forward graph to these variables and make the call to the forward graph. We use
call_with_info because we want to be able to retrieve the activations from the
forward graph and pass them to the backward graph (see
calling-a-subgraph)
The cross_entropy_with_grad operation returns the loss tensor and the gradient
to start backpropagation, which is 1 (dl/dl) unless you specify the additional
argument loss_scaling.
We create an output stream from the device to the host in order to retrieve loss values.
We call the backward graph and retrieve a dictionary mapping each tensor in the
forward graph to its corresponding gradient in the backward graph with
fwd_parent_ins_to_grad_parent_outs. This dictionary can then be used by the
optimiser to update the weights of the model.
Finally, we setup the properties for ir, specifying num_host_transfers, and
we return a popxl.Session so that we can execute our program.
Now we are ready to construct the model using the following hyper-parameters:
train_batch_size = 8
test_batch_size = 80
device = "ipu_hw"
learning_rate = 0.05
epochs = 1
training_data, test_data = get_mnist_data(test_batch_size, train_batch_size)
train_session, train_input_streams, train_variables, loss_stream = train_program(
train_batch_size, device, learning_rate
)
And train it as follows:
num_batches = len(training_data)
with train_session:
for epoch in range(1, epochs + 1):
print(f"Epoch {epoch}/{epochs}")
bar = tqdm(training_data, total=num_batches)
for data, labels in bar:
inputs: Mapping[popxl.HostToDeviceStream, np.ndarray] = dict(
zip(train_input_streams, [data.squeeze().float(), labels.int()])
)
loss = train_session.run(inputs)[loss_stream]
bar.set_description(f"Loss:{loss:0.4f}")
Epoch 1/1
Loss:0.0270: 100%|██████████| 7500/7500 [00:24<00:00, 306.92it/s]
After training we retrieve the trained weights to use them during inference and test the accuracy of the model.
To do that, we need to get the data stored in the tensor on the device with get_tensors_data.
train_vars_to_data = train_session.get_tensors_data(train_variables.tensors)
Validation
To test our model we need to create an inference-only program and run it on the test dataset.
def test_program(test_batch_size, device):
ir = popxl.Ir(replication=1)
with ir.main_graph:
# Inputs
in_stream = popxl.h2d_stream((test_batch_size, 28, 28), popxl.float32, "image")
in_t = ops.host_load(in_stream)
# Create graphs
facts, graph = Net().create_graph(in_t)
# Initialise variables
variables = facts.init()
# Forward
(outputs,) = graph.bind(variables).call(in_t)
out_stream = popxl.d2h_stream(outputs.shape, outputs.dtype, "outputs")
ops.host_store(out_stream, outputs)
ir.num_host_transfers = 1
return popxl.Session(ir, device), [in_stream], variables, out_stream
Again, we initialise the IR.
Then, we define the input stream, the graph and the factories for the variables.
We instantiate the variables and bind the graph to them, obtaining our bound graph.
Finally, we call the model. Since this time we do not need to retrieve
information from the call site, we can simply use call instead of
call_with_info.
We store the output in an output stream for later use.
We now call the function to create the test program and initialise the session:
test_session, test_input_streams, test_variables, out_stream = test_program(
test_batch_size, device
)
Before we run, we need to initialise the model variables with the weights we trained earlier.
To do that, once the test_session is created we can call
test_session.write_variables_data. This function requires a dictionary tensor_to_be_written : tensor_data_to_write.
It makes a call to test_session.write_variable_data(tensor,tensor_data) for
each tensor in the dictionary and then makes a single host to device transfer at
the end to send all data in one go.
In our case, tensor_to_be_written needs to be the test_session variables,
and tensor_data_to_write needs to be the tensor values, given as numpy arrays.
We already have a dictionary between train_session variables and their values,
the train_weights_data_dict retrieved with train_session.get_tensor_data.
We need a dictionary between the test_session variables and the
train_session variables, so that we can create the required test_variables : test_variables_data dictionary.
Provided test and training variables have the same names, we can use the
DotTree.to_mapping function to create this mapping. Given two DotTrees, for
common keys this function creates a dictionary of their values. So,
train_variables.to_mapping(test_variables) returns a dictionary popxl.Tensor : popxl.Tensor, where each key-value pair is made of a train variable and the
test variable with the same name.
With these two dictionaries, we can finally build the required test_variables : test_variables_data dictionary.
Earlier we saved the trained weights to train_vars_to_data.
# dictionary { train_session variables : test_session variables }
train_vars_to_test_vars = train_variables.to_mapping(test_variables)
# Create a dictionary { test_session variables : tensor data (numpy) }
test_vars_to_data = {
test_var: train_vars_to_data[train_var].copy()
for train_var, test_var in train_vars_to_test_vars.items()
}
We call .copy on each tensor because get_tensors_data returns a memory view
of the data. This may become invalid if the session is invalidated (or it may
change if we do something else later).
# Copy trained weights to the program, with a single host to device transfer
test_session.write_variables_data(test_vars_to_data)
# check that weights have been copied correctly
test_vars_to_data_after_write = test_session.get_tensors_data(test_variables.tensors)
for test_var, array in test_vars_to_data_after_write.items():
assert (array == test_vars_to_data[test_var]).all()
Finally, we can run the model and evaluate accuracy from the predictions of the model. The predictions do not need to be normalized.
def accuracy(predictions: np.ndarray, labels: np.ndarray):
ind = np.argmax(predictions, axis=-1).flatten()
labels = labels.detach().numpy().flatten()
return np.mean(ind == labels) * 100.0
num_batches = len(test_data)
sum_acc = 0.0
with test_session:
for data, labels in tqdm(test_data, total=num_batches):
inputs: Mapping[popxl.HostToDeviceStream, np.ndarray] = dict(
zip(test_input_streams, [data.squeeze().float(), labels.int()])
)
output = test_session.run(inputs)
sum_acc += accuracy(output[out_stream], labels)
test_set_accuracy = sum_acc / len(test_data)
print(f"Accuracy on test set: {test_set_accuracy:0.2f}%")
100%|██████████| 125/125 [00:01<00:00, 95.71it/s]Accuracy on test set: 96.16%
Conclusion
In this tutorial we explored the new PopXL API. We achieved the following:
built a simple Linear model (by subclassing
addons.Module) and ran it.created a multi-layer model with subgraphs and explored outlining. Outlining reduces program size by compiling subgraphs as callable functions rather than inlined code. We constructed subgraphs using
create_graph()and bound them explicitly to make 2 layers of a network.explored the DotTree syntax which is useful for accessing elements of the model and creating connections.
built and trained a full MNIST model. The gradients were calculated with the
addons.autodifftransform applied to the forward computational graph.
We will re-use the addons.Module class, in another tutorial, when we make a
custom optimiser.
To try out more features in PopXL look at our other tutorials.
You can also read our PopXL User Guide for more information.
As the PopXL API is still experimental, we would love to hear your feedback on it (support@graphcore.ai). Your input could help drive its future direction.
Generated:2022-07-22T13:03 Source:mnist.py SDK:2.6.0+1074 SST:0.0.7